Abstract
This is a short presentation of the light interception algorithm implemented in ARCHIMED-Phys, the model we use and develop in the FSPM team in the AMAP lab (CIRAD - Montpellier, France). It was presented at the FSPM 2020 conference (https://www.fspm2020.net/). Links:
- poster: https://www.researchgate.net/publicat...
- documentation of the software: https://archimed-platform.github.io/a...
- Github: https://github.com/ARCHIMED-platform
Abstract
Table of Contents
Introduction
Light modelling at the scale of organs is essential to account accurately for the complex interactions between biophysical processes such as photosynthesis, stomatal conductance and energy balance. Yet, the calculation of radiative exchanges at fine scales is computationally-intensive and it remains a hindrance to a widespread use of FSPMs despite advances in light modelling using either radiosity (Chelle and Andrieu, 1998) or raytracing (Bailey, 2018). This study shows that simplifications based on the discretization of radiative fluxes allow processing radiative exchanges in a natural environment while maintaining good accuracy on the simulation of biophysical processes such as carbon assimilation.
Material and Methods
The present study is based on biophysical simulations performed using the ARCHIMED model. Incident radiation is depicted as a set of specular fluxes (i.e. parallel rays) in discrete directions using the sun direction for direct radiation and predefined “turtle” directions for the diffuse radiation. The “turtle” directions are obtained by splitting the sky hemisphere into sectors of equal solid angle (Dauzat et al, 2001). Optionally, direct radiation can be distributed in neighboring “turtle” sectors (turtle only). For each direction, the scene is projected on an image plane and the interception of incident light is deduced from rasterized pixel projections. Additionally, Z-Buffering gives the overlay of scene objects and, in this regard, pixels can be viewed as rays traced from outside down to the ground level. Light scattering can thus be processed similarly to raytracing. In the case of Lambertian objects, we further assume that all rays scattered by an object carry the same energy whatever the “turtle” direction. Net assimilation (An) is calculated with Farquhar’s model (Farquhar et al. 1980), stomatal conductance with Medlyn’s model (Medlyn et al. 2011) and the leaf temperature is found by solving the energy balance of the system. Simulations are run on a dense three-dimensional scene including two palms (Elaeis guineensis) with the following configuration: latitude= 15°, Day of year 71, time steps of 30mn, clearness index Kt= 0.5. A “toricity” option is used to generate a virtually infinite canopy. The number of “turtle” directions is set to 6, 16, 46 or 136. The sun position is either integrated into the turtle or separately computed. The pixel density ranges from 341 to 6821 pixels m-2. The reference outputs are obtained with the highest number of directions and pixels. * Scene metrics: plot= 15.9m*9.2m, meshes= 24 863, triangles= 571 934, LAI= 3.2, leaflets= 24 493
Results and Discussions
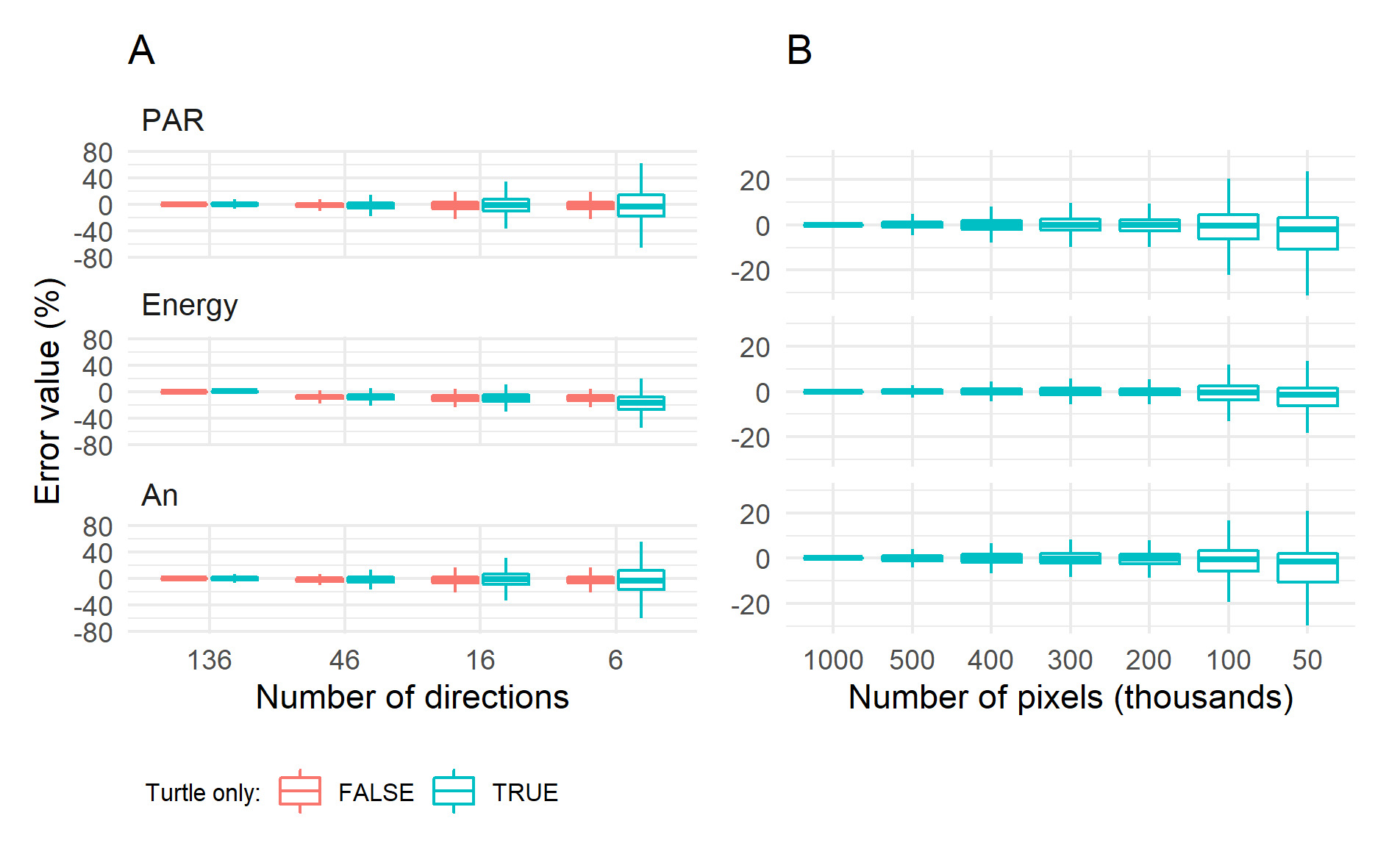
Figure 1 (left) illustrates the effect of the number of discrete light directions on the estimation of biophysical processes in comparison with the reference of 136 directions. Sampling the sun direction provides best results since direct radiation largely contributes to the PAR irradiance, the energy load of leaflets and, finally, their assimilation. Bias remain low when the sun direction is not sampled except when the number of “turtle” directions is decreased to six. The dispersion of residuals remains quite limited for 46 directions, meaning that reliable values can be obtained at leaflet scale for such configuration. Figure 1 (right) shows that a low pixel density (682 pixels m-2, i.e. 50 000 pixels) is sufficient to get a relatively unbiased estimation of carbon assimilation at plot level, but a higher density is necessary to get reliable estimation at leaflet scale. The reference configuration in the left pane of Fig. 1 generates 68.5M rays for each time step and, since several hits are recorded per ray (6 on average) this generates about 5 sub-rays that are used for the calculation of light scattering. Running the complete simulation with the reference configuration from the right pane of Fig. 1 lasts ~3.4 min for each time step (23M rays). This time can be decreased to only 2 seconds per step by storing partial scene illumination for each direction, but this preliminary step can be time-consuming, mainly during the multiple scattering for the PAR and NIR ranges. A considerable shortening is expected by treating light exchanges using directional form factors between pairs of objects instead of propagating scattered light by individual rays.
Conclusion
Using discrete ordinates allows performing accurate and unbiased simulations of light interception. Biases arise when decreasing the number of directions but with limited consequences on carbon assimilation. Larger biases occur when pixel density is too low to sample correctly individual leaflets. A configuration with 46 turtle directions for depicting both direct and diffuse radiation and a pixel density of 682 pixels m-2 allows fast computations while providing sufficient information to get precise light budget at fine scales.
References
Bailey, 2018, Ecological Modelling. 368:233-245, doi: 10.1016/j.ecolmodel.2017.11.022.
Chelle and Andrieu, 1998, Ecological Modelling 111:75-91, doi: 10.1016/S0304-3800(98)00100-8
Dauzat et al., 2001, Agric. & Forest Met. 109(2)143-160, doi: 10.1016/S0168-1923(01)00236-2
Farquhar et al., 1980, Planta. 149:78-90, doi: 10.1007/BF00386231
Medlyn et al., 2011, Global Change Biology. 17:2134-2144, doi: 10.1111/j.1365-2486.2010.02375.x